ORIGINAL ARTICLE
MELO, Nataly Sayonara da Silva [1], SILVA, Maria Goretti Varejão da [2], ALMEIDA, Anna Carolina Soares [3], MEDEIROS, Anna Karolyne de Araujo [4], SILVA, Daniel Dias da [5], SOUZA, Paula Mariana Salgueiro de [6], SILVA, Marcela Oliveira da [7], SOARES, Anísio Francisco [8], MENDONÇA, Marcelo [9], MEDEIROS, Elizabeth Sampaio de [10]
MELO, Nataly Sayonara da Silva, et al. Salmonella spp. virulent and resistant multidrug recovered from chicken carcasses in Brazil. Revista Científica Multidisciplinar Núcleo do Conhecimento. Year. 08, Ed. 04, Vol. 01, pp. 92-114. April 2023. ISSN: 2448-0959, Access link: https://www.nucleodoconhecimento.com.br/biology/salmonella-spp, DOI: 10.32749/nucleodoconhecimento.com.br/biology/salmonella-spp
ABSTRACT
The aim of this study was to evaluate the biofilm production, the susceptibility profile and the detection of resistance genes present in Salmonella spp isolates from fresh chicken carcasses sold in a Brazilian metropolis. From a total of 61 samples of fresh poultry carcasses, 21 were positive for the presence of Salmonella spp. Regarding the antimicrobial susceptibility test, (13/21) isolates tested were resistant to at least one antibiotic, corresponding to 61.9%, and 38% (08/21) were Resistant to Multiple Drugs. At least two resistance genes were identified in all isolates, especially the genes related to β-lactamases and Quinolones resistance. It was also observed that some Salmonella spp isolates showed identical genetic patterns. And all 21 isolates were able to form biofilm. The identification of Salmonella spp. biofilm forming and carrying different β-lactamase genes and determinants of resistance to quinolones demonstrates the capacity of these bacteria to accumulate various mechanisms of virulence and resistance to antimicrobials. Therefore, the spread of different clonal groups of Salmonella spp. MDR in poultry meat carcasses expressed in this attest to the need for effective controls to contain this microorganism, which besides being a risk to public health, is also responsible for considerable economic losses.
Keyword: Beta-lactams, Food poisoning, PCR, Public market.
INTRODUCTION
Salmonellosis is a foodborne disease caused by pathogenic Salmonella spp strains. Cases of Salmonellosis in humans occur mainly through the consumption of contaminated food or water. In most cases, food of animal origin, especially those from poultry, are the major sources of Salmonella spp. infections. The symptoms of the enteric infection are nausea, vomit, non-bloody diarrhea, fever, cold, abdominal pain, myalgia and headaches, and in immunodepressed patients it can lead to bacteremia, endocarditis, and death (GRIMONT and WEILL, 2007).
According to the Centres for diseases, control and prevention (CDC), 1.2 million cases of infections, 23.000 hospitalizations, and 450 deaths per year in the United States of America are caused by Salmonella spp, approximately. And the infections related to the ingestion of contaminated food represents 1.0 million of the cases, approximately (CDC, 2020). In Brazil, according to the Ministry of Health, a total of 12.660 cases of foodborne illness was reported between 2000 and 2017, and Salmonella spp. was one of the major agents reported in these cases, which represents a total of 35% of the cases reported (BRASIL, 2019).
In most salmonellosis cases, healthy individuals do not need treatment. However, immunodepressed patients need to go through treatment (SERENO et al., 2017). Nevertheless, the indiscriminate use of antimicrobials, especially in animal production has led to an increased number of microorganisms resistant to therapeutic agents, leading to a limited number of antimicrobial options, which can result in treatment failures (ALEKSHUN and LEVY, 2007; PENG et al., 2014). Over the years, there has been an increase in the prevalence of Salmonella strains multiple drugs resistant, mainly in poultry products, such as poultry meat (AZEVEDO, 2014; BAPTISTA et al., 2018; DUARTE et al., 2009; FITCH et al., 2016; BONI et al., 2011; REZENDE et al., 2005; RIBEIRO et al., 2007; THRELFALL, 2002).
Therefore, the emergence of multidrug-resistant Salmonella (MDR) is a worldwide concern, due to the increased hospitalization rates and death. This phenomenon is a consequence of the extensive use of antibiotics by humans, and also in animal production.
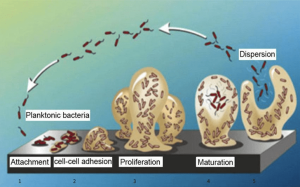
Moreover, Salmonella species have the ability to form biofilm, where cells microorganisms are embedded in an extracellular matrix, so these microorganisms can adhere to biotic or abiotic surfaces (DAVEY and O’TOOLE, 2000; FLEMMING et al., 2016). The complex matrices of the biofilm keep the microbial cells protected from the action of sanitation process and antimicrobial agents, therefore, when pathogenic microorganisms are involved in the biofilm, there is a huge risk to contaminate the food, which leads to a public health problem. (KASNOWSKY et al., 2010).
Hence, the aim of this study was to characterize the determinants of antibiotic resistance, and to evaluate the production of biofilm in Salmonella spp. strains recovered from fresh poultry meat sold in a Brazilian metropolis.
METHODS
Sampling and identification of the poultry carcasses
A total of 61 poultry meat carcasses were purchased from 07 different public markets in the city of Recife in the state of Pernambuco, Brazil, between 2018 to 2019 (figure 1 and table 1). The samples were sent to the Meat and Milk Inspection Laboratory (LICAL), of the Department of Veterinary Medicine in the Federal Rural University of Pernambuco (UFRPE).
At the laboratory, the surface of each poultry carcasses pack was wiped with 70 percent alcohol. Then, 25g of the carcases were randomly taken and placed in individual sterile stomacher bags with 225 ml of Buffered Peptone Water (Kasvi, Brazil), and homogenized in stomacher for 2 min (Kasvi, Brazil) and incubated at 37 °C for 24 hours for the pre-enrichment stage. For the enrichment stage, 0.1 ml of the pre-enriched broth was transferred into 10 ml of Tetrathionate broth (Merck e pais) and then incubated at 41.5 °C for 24 hours, and 1.0 ml of the pre-enriched broth was also transferred into 10 ml of Rappaport and incubated at 37 °C for 24 hours for selective enrichment. After the incubation, one loopful of the TT broth and Rappaport cultures were streaked onto Xylose Lysine Agar (XLD) (Kasvi, Brazil) and Hektoen Enteric Agar (HE) (Kasvi, Brazil) plates and incubated at 37 °C for 24h. Three presumptive Salmonella colonies on the plates were selected and streaked onto Nutrient agar (Merck, pais-) and incubated at 37.8 °C for 24 hours. Then, colonies from the Nutrient agar were submitted to biochemical and serological tests (ISO, 657912017).
Figure 1: Public markets where the samples were acquired
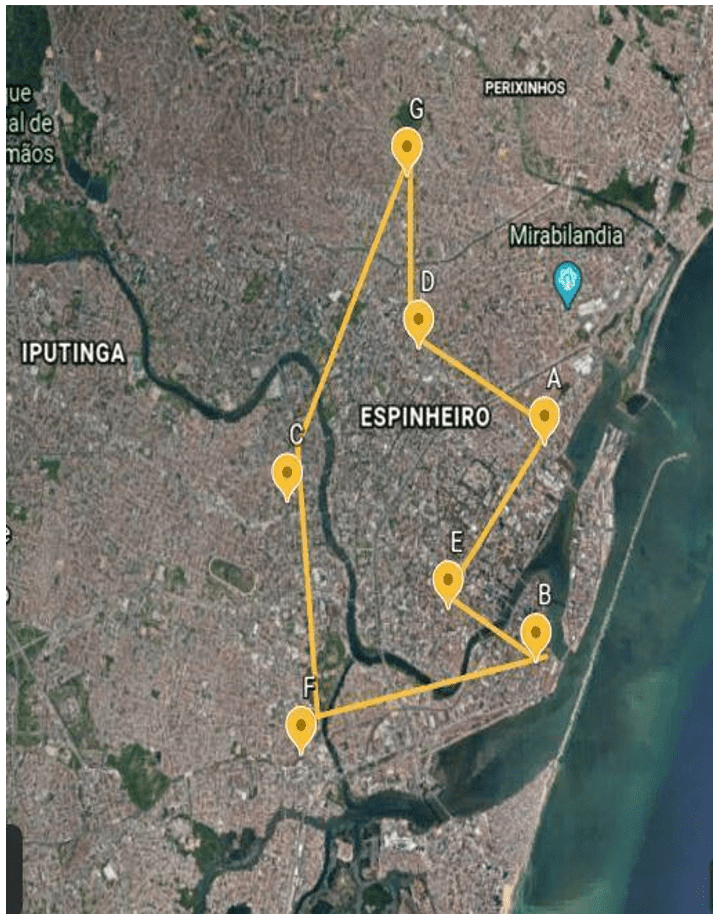
Table 1: Number of samples acquired in different public markets in the city of Recife – Pernambuco, Brazil
| Public Market | Samples acquired |
| A | 1, 2, 3 22, 23, 24, 25, 26 |
| B | 4, 5, 6, 7, 8, 9, 27, 28, 29 |
| C | 30, 31, 32 |
| D | 10, 33, 34, 35, 36, |
| E | 11, 37, 38, 39, 40, 41 |
| F | 12, 13, 14, 15, 16, 17, 18, 45, 46,47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61 |
| G | 18, 20, 21, 42, 43, 44 |
Source: Authors.
Susceptibility profile to antimicrobials
The susceptibility profile test was done by the agar disc-diffusion method with the following antimicrobial agents: ampicillin, chloramphenicol, ciprofloxacin, ceftriaxone, sulfamethoxazole-trimethoprim, imipenem, amoxicillin-clavulanic acid, ceftazidime, cefotaxime and aztreonam, according to the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2018).
Detection of resistance genes
The DNA used in the molecular analyses was obtained from bacterial suspensions made up of fresh colonies, which were previously grown for 16-18 hours in Luria Bertani agar and inoculated in approximately 300 μL ultrapure water free of nuclease homogenized with the aid of a vortex tube shaker (Vision Scientific).
The investigation of genes to β-lactams and quinolones resistance was performed by PCR. The thermal cycling conditions were those described by the authors listed in the primer table.
Table 2: Primers used to detect the β-lactamases genes
| Primer | 5’ 3’ Sequences | Target Gene | Product | References | |
| KPC | F | TGTCACTGTATCGCCGTC | blaKPC | 1011pb | (Bratu et al., 2005) |
| R | CTCAGTGCTCTACAGAAAACC | ||||
| CTX-M | F | SCSATGTGCAGYACCAGTAA | blaCTXM | 500pb | (Cao et al., 2002) |
| R | CCGCRATATGRTTGGTGGTG | ||||
| TEM | F | TCGGGGAAATGTGCGCG | blaTEM | 700pb | |
| R | TGCTTAATCAGTGAGGCACC | ||||
| SHV | F | TTATCTCCCTGTTAGCCACC | blaSHV | 900pb | |
| R | GATTTGCTGATTTCGCTCGG | ||||
| R | AAGCAGACTTGACCTGA | ||||
| qnrA | F | AGA GGA TTT CTC ACG CCA GG | qnrA | 580pb | (Cattoir et al., 2007). |
| R | TGC CAG GCA CAG ATC TTG AC | ||||
| qnrB | F | GGM ATH GAA ATT CGC CAC T | qnrB | 264pb | |
| R | TTT GCY GYY CGC CAG TCG AA | ||||
| qnrC | F | GGG TTG TAC ATT TAT TGA ATC | qnrC | 447pb | (Wang et al., 2009) |
| R | TCC ACT TTA CGA GGT TCT | ||||
| qnrD | F | CGA GAT CAA TTT ACG GGG AAT A | qnrD | 582pb | (Cavaco et al., 2009). |
Source: Authors.
Analysis of the genetic profile of the isolates
The comparison and identification of genetic variations in the content of bacterial strains was performed by ERIC-PCR. The primers used for the reaction were ERIC-1R (5′ ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC2 (5’AAGTAAGTGACTGGGGGGGCCG-3′) (Sigma Aldrich) according to (VERSALOVIC et al., 1991), with modification described by (FENDRI et al., 2013).
Biofilm production capacity
The biofilm forming capacity was evaluated according to the methodology proposed by (STEPANOVIĆ et al., 2000; STEPANOVIĆ et al., 2007). Each strain was diluted to 108CFU/mL (0.5 in the MacFarland scale) using Trypticase Soy Broth (TSB) (Merck), and 200μL were cultured in three wells of the 96-well flat-bottom polystyrene microplate (Nest®). A total of 69 wells were used to test 21 strains; the other 3 wells received the Salmonella Typhimurim ATCC 14028 as positive control, and 3 wells received the negative control (non-inoculated culture medium). The plates containing Salmonella spp. strains and controls were incubated at 35ºC for 96 hours. The plate was washed three times with phosphate buffered saline (PBS pH 7.2) and stained with crystal violet at 1% for 15 minutes. After washing three times with distilled water and drying at room temperature, absorbance was read in a Polaris (Celer®) microplate reader at 492 nm wavelength (SERENO et al., 2017).
The optical density (OD) of each Salmonella spp. strain was obtained by the arithmetic mean of the absorbance of three wells and this value was compared with the mean absorbance of negative controls (ODnc). After that, the strains were classified in no biofilm producer (OD≤ODnc); weak biofilm producer (ODnc<ODs≤2.ODnc); moderate biofilm producer (2.ODnc<ODs≤4.ODnc); and strong biofilm production (4.ODnc<ODs) classification given according to (STEPANOVIĆ et al., 200; STEPANOVIĆ et al., 2007).
RESULTS
Prevalence of Salmonella spp. in poultry meat carcass samples
From the 61 samples of poultry meat carcasses collected in the public markets, 34% (21) were contaminated by Salmonella spp. The prevalence of Salmonella spp. on each public market is expressed in figure 3, and the data is detailed in table 3.
Figure 2: Prevalence of Salmonella spp. isolates from each public market in the city of Recife – Pernambuco, Brazil
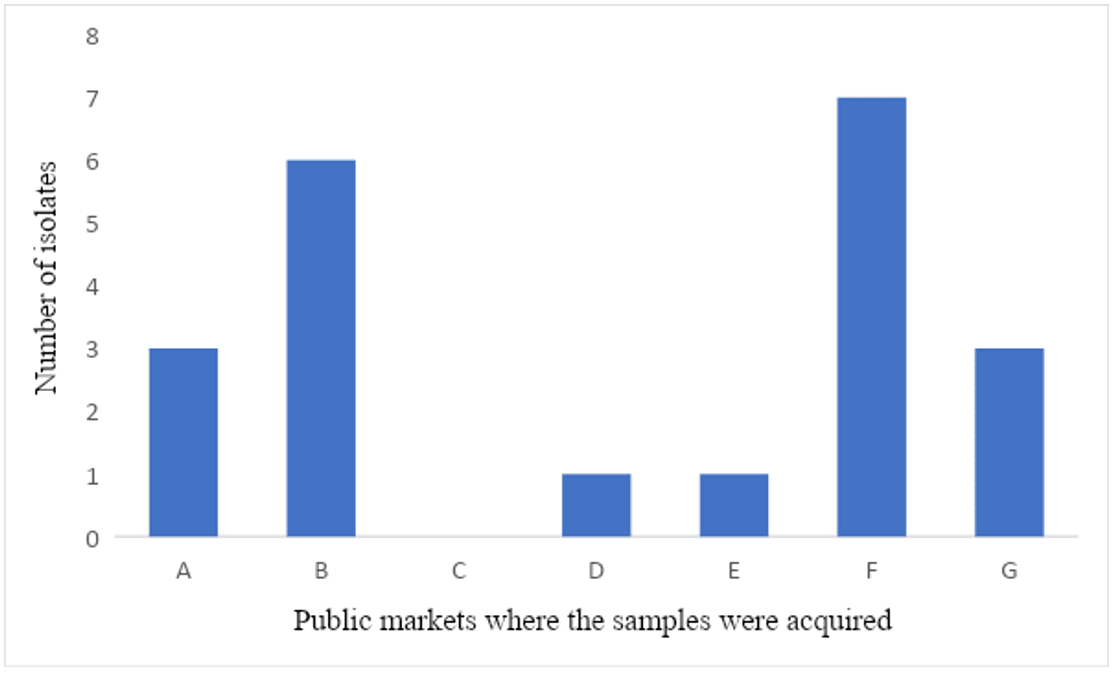
Table 3: Salmonella spp. isolates from each public market in the city of Recife – Pernambuco, Brazil
| Isolates | Public market | Year |
| 1 | A
|
2018 |
| 2 | 2019 | |
| 3 | 2019 | |
| 4 | B | 2018 |
| 5 | 2018 | |
| 6 | 2018 | |
| 7 | 2018 | |
| 8 | 2018 | |
| 9 | 2018 | |
| 10 | D | 2019 |
| 11 | E | 2019 |
| 12 | F
|
2019 |
| 13 | 2019 | |
| 14 | 2019 | |
| 15 | 2019 | |
| 16 | 2019 | |
| 17 | 2019 | |
| 18 | 2019 | |
| 19 | G
|
2018 |
| 20 | 2019 | |
| 21 | 2019 |
Source: Authors.
Figure 3: Antimicrobial resistance to individual antimicrobial agents among Salmonella spp. isolates from poultry carcasses sold in public markets in the city of Recife – Brazil
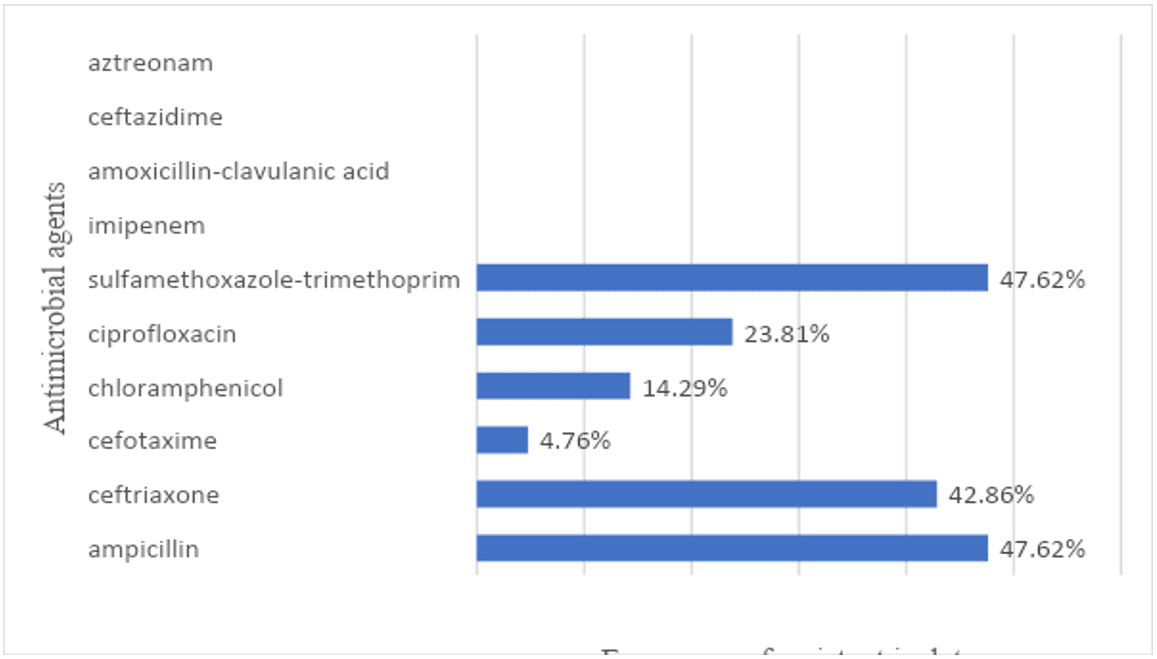
Susceptibility perfil
From the 21 isolates, 13 (61,9%) were resistant to at least one antimicrobial agent. The antimicrobial resistance to individual antimicrobial agents among Salmonella isolates is expressed in figure.3. The detailed data from the antimicrobial susceptibility test is expressed in table 4.
Table 4. Antimicrobial susceptibility profile of Salmonella spp. isolates from chicken carcasses acquired in public markets in the city of Recife – PE, between 2018 and 2019
| Isolate | AMP | AMC | CRO | CTX | CAZ | IPM | ATM | CLO | CIP | SUT |
| 1 | R | S | R | S | S | S | S | S | R | R |
| 2 | S | S | S | S | S | S | S | S | I | S |
| 3 | S | S | S | S | S | S | S | S | R | R |
| 4 | R | S | I | S | S | S | S | S | S | S |
| 5 | S | S | S | S | S | S | S | S | I | S |
| 6 | S | S | I | S | S | S | S | S | R | S |
| 7 | S | S | I | S | S | S | S | S | I | S |
| 8 | S | S | S | S | S | S | S | S | R | S |
| 9 | S | S | I | S | S | S | S | S | I | S |
| 10 | S | S | S | S | S | S | S | S | I | S |
| 11 | S | S | S | S | S | S | S | S | I | S |
| 12 | R | S | R | S | S | S | S | S | I | R |
| 13 | R | S | R | S | S | S | S | S | I | R |
| 14 | S | S | S | S | S | S | S | S | I | S |
| 15 | R | S | R | S | S | S | S | R | S | R |
| 16 | R | S | R | S | S | S | S | R | S | R |
| 17 | R | S | R | S | S | S | S | R | S | R |
| 18 | R | S | R | S | S | S | S | S | S | R |
| 19 | S | S | S | S | S | S | S | S | S | S |
| 20 | R | S | R | S | S | S | S | S | S | R |
| 21 | R | S | R | R | S | S | S | S | R | R |
AMP – Ampicillin, CLO – Chloramphenicol, CIP – Ciprofloxacin, CRO- Ceftriaxone, SUT – Sulfamethoxazole-trimethoprim, IPM – Imipenem, AMC – amoxicillin-clavulanic acid, CAZ – Ceftazidime, CTX- Cefotaxime and ATM – Aztreonam, S – Sensitive; R- Resistant; I-. Intermediate. Source: Authors.
At least two resistance genes were identified in all isolates, among which the presence of resistance genes encoding β-lactamases (bla) and resistance genes encoding quinolones (qnr) were observed in the molecular tests carried out in the present study, and the results are detailed in table 3.
Table 5: Resistance genes detected in each Salmonella spp isolate, recovered from poultry carcasses acquired in public markets in the city of Recife – Pernambuco between the years 2018 and 2019
| β-lactams resistant | Quinolones resistants | |||||||
| Isolates | BlaTEM-like | BlaSHV-like | BlaCTX-M-like | BlaKPC-2 | qnrA | qnrB | qnrC | qnrD |
| 1 | + | – | – | – | + | + | – | – |
| 2 | + | – | – | – | + | + | – | – |
| 3 | + | – | – | – | + | + | – | + |
| 4 | + | – | – | – | + | + | – | – |
| 5 | + | – | – | – | + | + | – | + |
| 6 | + | – | – | – | + | + | – | + |
| 7 | + | – | – | – | + | + | – | + |
| 8 | + | – | – | – | + | + | – | + |
| 9 | + | – | – | – | + | + | – | + |
| 10 | + | – | + | – | – | + | – | – |
| 11 | + | – | – | – | – | + | – | – |
| 12 | + | – | + | – | – | + | – | – |
| 13 | + | – | + | – | – | + | – | – |
| 14 | + | – | + | – | – | + | – | – |
| 15 | + | – | + | – | – | + | – | – |
| 16 | + | – | + | – | – | + | – | + |
| 17 | – | – | + | – | – | + | – | + |
| 18 | + | – | + | – | – | + | – | + |
| 19 | + | – | – | – | – | + | – | + |
| 20 | + | – | – | – | – | + | – | + |
| 21 | + | – | – | – | + | + | – | + |
Source: Authors.
Clonal relationship
The isolates showed a pattern ranging from 3 to 9 strips, which revealed the presence of different genotypes pattern indistinguishable from each other and others whose standard is different for only one or two bands. Some isolates of Salmonella spp. showed identical genetic patterns (2, 3, 4, 5, 6 and 7). Samples 9, 11, 19, 20 and 21 were closely related genetically, as they have a similar pattern of strips, differing only in one (9, 21) or two strips (11 and 20).
Biofilm forming
All the 21 Salmonella spp. isolates tested were biofilm producers, and these isolates were classified as weak biofilm producers according to (STEPANOVIĆ et al., 2000; STEPANOVIĆ et al., 2007).
DISCUSSION
This work identified Salmonella spp. contaminating chicken carcasses sold in public markets in a Brazilian metropolis. In addition, we detected the resistance mechanisms present in these bacterial isolates and confirmed that they were all biofilm producers. The results that are presented here showed several isolates with a genetic proximity and revealed that the others are clones.
Isolates 2, 3, 4 and 5 have an identical genetic profile, although they were collected in two different markets, Market A and Market B, which are about 5 km apart from each other. This finding indicates that the sellers may have the same supplier, where the contamination by Salmonella MDR started. Market F presented a greater number of strains characterised as clones (12, 14, 15, 16 and 18). In addition to the possibility of the same supplier, according to a set of failures in hygienic conditions, such as the exposure of products to dust and insects and the inappropriate sharing of utensils such as knives and containers between traders, they can contribute to the contamination of the carcasses, which results in the dissemination of isogenic isolates (AZEVEDO, 2014).
The presence of Salmonella spp. in poultry meat was also observed in the studies done by (BAPTISTA et al., 2018; MELO et al., 2020; YIN et al., 2021). However, our findings differ from these studies due to the higher prevalence of Salmonella spp isolates.
The contamination by Salmonella spp. detected in the samples may have been due to previous contamination in the slaughterhouse itself, because of failures in the process of slaughtering (LOPES et al., 2007; VON RÜCKERT et al., 2009). It was also observed that traders removed the carcasses from the original packaging and placed them in plastic bowls and later sold them at room temperature (88 ºF), which is considered a fraud. This temperature promotes the multiplication of Salmonella spp., as these microorganisms can multiply at temperatures around 5°C to 45°C (MAHMOUD, 2012) This problem related to the temperature was also noticed in the studies done by (KHAN et al., 2018; JARQUIN et al., 2015).
Therefore, this practice increases the risk of product contamination, mainly due to inadequate handling practices and inadequate sanitary hygienic conditions and to what is observed in these public markets, as it was found that tables and benches were not cleaned in 59% (36/61) of the places. Regarding the handlers, it was observed that 100% did not have their nails well cut and clean. In addition, the same employee who took care of the meat was the same one in the checkout counter. Consequently, these flaws in the processes of good handling practices observed in these markets can contribute to the contamination and spread of Salmonella spp. in this product. This transference can occur between various interactions between man, food and the environment. Therefore, cross-contamination is the basis for the spread of Salmonella spp.
From the results from the susceptibility test against antimicrobials agents in the present study, it was observed that 38% (08/21) of the Salmonella spp strains were Resistant to Multiple Drugs (MDR), that is, bacteria that are resistant to at least three classes of antimicrobial agents (MAGIORAKOS et al., 2011).
The occurrence of Salmonella spp strains resistant to multiple antimicrobials in poultry meat in Brazil is a growing reality, due to the data reported in the present study, and other researches done by (ZIECH, 2015; BAPTISTA et al., 2018). It might happen because of the intensive use of antimicrobial agents in the animal production to treat the animals, and also to improve the production.
The greatest implication regarding the dissemination of multidrug resistant zoonotic strains is related to therapeutic failures in the face of several conditions, both in human and animal cases. In addition, this reality leads to losses in animal herds, since when present, these resistant strains are difficult to eliminate. The permanence of pathogenic and multidrug resistant strains in the poultry production is even more worrying, since the animal density is higher, and consequently, there is a greater spread of the microorganisms in a shorter period of time.
It was observed that only 4.28% (03/21) of the isolates were resistant to Chloramphenicol. This result may reflect the ban on the use of this agent as a therapeutic agent and growth promoter in animal production in Brazil (PACHECO-SILVA et al., 2014).
On the other hand, 42.85% (09/21) of the isolates were resistant to Ceftriaxone, 3rd generation cephalosporin. The use of this drug in animal production is prohibited in Brazil (MION et al., 2014). Therefore, this result indicates that this drug or others from the same antimicrobial class is still being used in animal production in Brazil.
In addition, 42.85 (9/21) of the isolates were resistant to three or more antimicrobial drugs. Then, when comparing the resistant profile with the markets, it was noticed that isolates number 13, 15, 16, 17, 18, which were resistant to up to three drugs were from the same public market (public market F). It suggests that there may have been cross contamination at the market, or that these chicken carcasses may have suffered previous contamination in the poultry slaughterhouse.
Hence, the occurrence of Salmonella MDR strains in food animal origin is increasing in Brazil (BAPTISTA et al., 2018; MELO et al., 2020; VOSS-RECH et al., 2015). Infections related to multidrug resistant strains are associated with high morbidity and mortality, compared to the sensitive ones, since these microorganisms represent a barrier in the treatment of human and animal diseases.
The presence of such resistance genes detected in Salmonella spp. isolates from food of animal origin represents a serious threat to public health, since the horizontal transmission of resistance genes occurs mainly by plasmids encoding β-lactamases (GYLES, 2008). In addition, these genes are considered from community origin, and are common in hospital environments (SUN et al., 2013). Thus, the dissemination of resistant strains points out the importance of the Salmonella spp control in poultry meat (BORGES et al., 2019; SIVASANKAR et al., 2020).
The presence of biofilm-forming strains of Salmonella spp. observed in the present study points out the risk that this finding may bring to the food industry and the public health. The ability to form biofilm protected the microorganisms inside the biofilm, and thus less susceptible to external factors, such as the action of antimicrobial agents (ZIECH, 2015; SERENO et al., 2017; BORGES et al., 2018).
In addition, biofilms promote the permanence of pathogenic and spoilage microorganisms, which cause illnesses to consumers, and the depreciation of the final product due to physical-chemical and sensory changes (KASNOWSKY et al., 2010; SINGH et al., 2017).
Finally, this investigation showed that the identification of Salmonella spp. producer of biofilm and carrying different β-lactamase genes and determinants of resistance to quinolones demonstrates the ability of these bacteria to accumulate various mechanisms of virulence and resistance to antimicrobials. The selective pressure is exerted by the indiscriminate use of antibiotics in agriculture is an important factor for the acquisition of different mechanisms of resistance and dissemination of these strains. Furthermore, the spread of different clonal groups of Salmonella spp. MDR, in chicken carcasses, shown in this study attests to the need for effective controls to contain this microorganism, which besides being a risk to public health, is also responsible for considerable economic losses.
FINANCIAL SUPPORT
The authors thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for the scholarship provided.
REFERENCES
ALEKSHUN, M. N.; LEVY, S. B. Molecular Mechanisms of Antibacterial Multidrug Resistance. Cell, v. 128, n. 6, p. 1037–1050, 23 mar. 2007. Available from: https://doi.org/10.1016/j.cell.2007.03.004. Acessed in: 02 mar. 2021.
AZEVEDO, S. M. M. Farmacologia dos antibióticos beta-lactâmicos. Porto, Portugal: Universidade Fernando Pessoa, 2014. Available from: https://bdigital.ufp.pt/handle/10284/4412. Acessed in: 02 mar. 2021.
BAPTISTA, D. Q. et al. Prevalência e susceptibilidade antimicrobiana de sorotipos de Salmonella spp. isolados de frangos vivos e carcaças no estado do Rio de Janeiro. Pesquisa Veterinária Brasileira, v. 38, n. 7, p. 1278–1285, jul. 2018. Available from: https://doi.org/10.1590/1678-5150-PVB-5289. Acessed in: 02 mar. 2021.
BORGES, K. A. et al. Biofilm formation capacity of Salmonella serotypes at different temperature conditions. Pesquisa Veterinária Brasileira, v. 38, n. 1, p. 71–76, jan. 2018. Available from: https://doi.org/10.1590/1678-5150-pvb-4928. Acessed in: 02 mar. 2021.
BORGES, K. A.; MARTELO, E. B.; DOS SANTOS, L. A.; FURIAN, T. Q.; CISCO, I. C.; MANTO, L.; DOS SANTOS, L. R. Detection and quantification of Salmonella spp. in poultry slaughterhouses of southern Brazil. The Journal of Infection in Developing Countries, [S. l.], v. 13, n. 05, p. 455–460, 2019. DOI: 10.3855/jidc.11107. Available from: https://jidc.org/index.php/journal/article/view/32053516. Acessed in: 02 mar. 2021.
BRATU, S.; LANDMAN, D.; ALAM, M.; TOLENTINO, E.; QUALE, J. Detection of KPC Carbapenem-Hydrolyzing Enzymes in Enterobacter spp. from Brooklyn, New York. Antimicrobial Agents and Chemotherapy, v. 49, n. 2, p. 776–778, fev. 2005. Available from: https://doi.org/10.1128/AAC.49.2.776-778.2005. Acessed in: 02 mar. 2021.
BRASIL. Manual Integrado de Vigilância, Prevenção e Controle de Doenças Transmitidas por Alimentos. Ministério da Saúde, Secretária de Vigilância em Saúde, 2019. Available from: http://portalarquivos2.saude.gov.br/images/pdf/2019/fevereiro/15/Apresenta oSurtos-DTA—Fevereiro-2019.pdf. Acessed in: 13 Aug. 2019.
BONI, H. F. K.; CARRIJO, A. S.; FASCINA, V. B. Detection of Salmonella spp. in broiler buildings and stuff of slaughterhouse in the central region of Mato Grosso do Sul. R. bras. Saúde Prod. Anim., 2011. Available from: http://revistas.ufba.br/index.php/rbspa/article/view/1949. Acessed in: 02 mar. 2021.
CAO, V.; LAMBERT, T.; NHU, D. Q.; LOAN, H. K.; HOANG, N. K.; ARLET, G.; COURVALIN, P. Distribution of Extended-Spectrum β-Lactamases in Clinical Isolates of Enterobacteriaceae in Vietnam. Antimicrobial Agents and Chemotherapy, v. 46, n. 12, p. 3739–3743, dez. 2002. Available from: https://doi.org/10.1128/AAC.46.12.3739-3743.2002. Acessed in: 02 mar. 2021.
CATTOIR, V.; POIREL, L.; ROTIMI, V.; SOUSSY, C. J.; NORDMANN, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. Journal of Antimicrobial Chemotherapy, v. 60, n. 2, p. 394–397, 1 ago. 2007. Available from: https://doi.org/10.1093/jac/dkm204. Acessed in: 02 mar. 2021.
CAVACO, L. M.; HASMAN, H.; XIA, S.; AARESTRUP, F. M. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrobial Agents and Chemotherapy, v. 53, n. 2, p. 603–608, fev. 2009. Available from: https://doi.org/10.1128/AAC.00997-08. Acessed in: 02 mar. 2021.
CENTRES FOR DISEASES, CONTROL AND PREVENTION – CDC. Salmonella. Centres For Diseases, Control And Prevention, 2020. Available from: https://www.cdc.gov/salmonella/index.html. Acessed in: 02 mar. 2021.
DAVEY, M. E.; O’TOOLE, G. A. Microbial Biofilms: from Ecology to Molecular Genetics. Microbiology and Molecular Biology Reviews, v. 64, n. 4, p. 847–867, dez. 2000. Available from: https://doi.org/10.1128/mmbr.64.4.847-867.2000. Acessed in: 02 mar. 2021.
DUARTE, D. A. M. et al. Occurrence of Salmonella spp. in broiler chicken carcasses and their susceptibility to antimicrobial agents. Brazilian Journal of Microbiology, v. 40, n. 3, p. 569–573, set. 2009. Available from: https://doi.org/10.1590/S1517-83822009000300020. Acessed in: 02 mar. 2021.
FENDRI, I.; BEN HASSENA, A.; GROSSET, N.; BARKALLAH, M.; KHANNOUS, L.; CHUAT, V.; GAUTIER, M.; GDOURA, R. Genetic Diversity of Food-Isolated Salmonella Strains through Pulsed Field Gel Electrophoresis (PFGE) and Enterobacterial Repetitive Intergenic Consensus (ERIC-PCR). PLOS ONE, v. 8, n. 12, p. e81315, 3 dez. 2013. Available from: https://doi.org/10.1371/journal.pone.0081315. Acessed in: 02 mar. 2021.
FITCH, F. M.; CARMO-RODRIGUES, M. S.; OLIVEIRA, V. G. S.; GASPARI, M. V.; DOS SANTOS, A.; DE FREITAS, J. B.; PIGNATARI, A. C. C. β-Lactam Resistance Genes: Characterization, Epidemiology, and First Detection of blaCTX-M-1 and blaCTX-M-14 in Salmonella spp. Isolated from Poultry in Brazil—Brazil Ministry of Agriculture’s Pathogen Reduction Program. Microbial Drug Resistance, v. 22, n. 2, p. 164–171, 8 mar. 2016. Available from: https://doi.org/10.1089/mdr.2015.0143. Acessed in: 02 mar. 2021.
FLEMMING, H. C.; WINGENDER, J.; SZEWZYK, U.; STEINBERG, P.; RICE, S. A.; KJELLEBERG, S. Biofilms: an emergent form of bacterial life. Nature Reviews Microbiology, v. 14, n. 9, p. 563–575, 11 ago. 2016. Available from: https://doi.org/10.1038/nrmicro.2016.94. Acessed in: 02 mar. 2021.
GRIMONT, P.; WEILL, F. X. Antigenic formulae of the Salmonella servovars: WHO Collaborating Centre for Reference and Research on Salmonella. 9th Ed. Inst. Pasteur, 2007. 1–166 p.
GYLES, C. L. Antimicrobial resistance in selected bacteria from poultry. Animal Health Research Reviews, v. 9, n. 2, p. 149–158, 2008. Available from: https://doi.org/10.1017/S14662523080015522. Acessed in: 02 mar. 2021.
JARQUIN, C.; ALVAREZ, D.; MORALES, O.; MORALES, A. J.; LÓPEZ, B.; DONADO, P.; VALENCIA, M. F.; ARÉVALO, A.; MUÑOZ, F.; WALLS, I.; DOYLE, M. P.; ALALI, W. Q. Salmonella on Raw Poultry in Retail Markets in Guatemala: Levels, Antibiotic Susceptibility, and Serovar Distribution. Journal of Food Protection, v. 78, n. 9, p. 1642–1650, set. 2015. Available from: https://doi.org/10.4315/0362-028X.JFP-15-117. Acessed in: 02 mar. 2021.
KASNOWSKY, M. C. et al. Formação de biofilme na indústria de alimentos e métodos de validação de superfícies. Revista Científica Eletrônica de Medicina Veterinária, v. 15, p. 1679–7353, jul. 2010. Available from: http://faef.revista.inf.br/imagens_arquivos/arquivos_destaque/fxPTiYWerLkT9Si_2013-6-25-16-32-0.pdf. Acessed in: 02 mar. 2021.
KHAN, A. S.; GEORGES, K.; RAHAMAN, S.; ABDELA, W.; ADESIYUN, A. A. Prevalence and serotypes of Salmonella spp. on chickens sold at retail outlets in Trinidad. PLOS ONE, v. 13, n. 8, p. e0202108, ago. 2018. Available from: https://doi.org/10.1371/journal.pone.0202108. Acessed in: 02 mar. 2021.
LOPES, M.; GALHARDO, J. A.; DE OLIVEIRA, T.; TAMANINI, R.; SANCHES, F.; MULLER, E. Pesquisa de Salmonella spp. e microrganismos indicadores em carcaças de frango e água de tanques de pré-resfriamento em abatedouro de aves. Semina: Ciências Agrárias, v. 28, n. 3, p. 465–476, 30 ago. 2007. Available from: https://doi.org/10.5433/1679-0359.2007v28n3p465. Acessed in: 02 mar. 2021.
MAGIORAKOS, A. P.; SRINIVASAN, A.; CAREY, R. B.; CARMELI, Y.; FALAGAS, M. E.; GISKE, C. G.; HARBARTH, S.; HINDLER, J. F.; KAHLMETER, G.; OLSSON-LILJEQUIST, B.; PATERSON, D. L.; RICE, L. B.; STELLING, J.; STRUELENS, M. J.; VATOPOULOS, A.; WEBER, J. T.; MONNET, D. L. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases, v. 18, n. 3, p. 268–281, 2012. Available from: https://pubmed.ncbi.nlm.nih.gov/21793988/. Acessed in: 02 mar. 2021.
MAHMOUD, B. S. M. Salmonella – A Dangerous Foodborne Pathogen, Nursing times, 2012. Available from: https://doi.org/10.1049/cm.1989.0039. Acessed in: 02 mar. 2021.
MELO, R. T. et al. Salmonella Minnesota de origem avícola apresenta fatores de virulência e risco potencial aos humanos. Arquivo Brasileiro de Medicina Veterinária e Zootecnia, v. 72, n. 4, p. 1353–1362, jul. 2020. Available from: https://doi.org/10.1590/1678-4162-10884. Acessed in: 02 mar. 2021.
MION, L.; COLLA, F. L.; CISCO, I. C.; WEBBER, B.; DIEDRICH, L. N.; PILOTTO, F.; RODRIGUES, L. B.; NASCIMENTO, V. P. do; SANTOS, L. R. dos. Perfil de resistência a antimicrobianos por Salmonella Heidelberg isoladas de abatedouro avícola em 2005 e 2009. Acta Scientiae Veterinariae, v. 42, n. 1, p. 1–5, 2014. Available from: https://www.redalyc.org/articulo.oa?id=289029240017. Acessed in: 02 mar. 2021.
PACHECO-SILVA, É.; SOUZA, J. R. de; CALDAS, E. D. Resíduos de medicamentos veterinários em leite e ovos. Química Nova, v. 37, n. 1, p. 111–122, 2014. Available from: https://doi.org/10.1590/S0100-40422014000100020. Acessed in: 02 mar. 2021.
PENG, M.; SALAHEEN, S.; BISWAS, D. Animal Health: Global Antibiotic Issues. Encyclopedia of Agriculture and Food Systems, p. 346–357, jan. 2014. Available from: https://doi.org/10.1016/B978-0-444-52512-3.00187-X. Acessed in: 02 mar. 2021.
REZENDE, C. S. M. e; MESQUITA, A. J. de; ANDRADE, M. A.; LINHARES, G. F. C.; MESQUITA, A. Q. de; MINAFRA, C. S. Sorovares de Salmonella isolados de carcaças de frangos de corte abatidos no Estado de Goiás, Brasil, e perfil de resistência a antimicrobianos. Revista Portuguesa de Ciências Veterinárias, Lisboa, v. 100, n. 555-556, p. 199-203, jul./dez. 2005. Available from: http://repositorio.bc.ufg.br/handle/ri/12309. Acessed in: 02 mar. 2021.
RIBEIRO, A. R. et al. Salmonella spp. in raw br
oiler parts: occurrence, antimicrobial resistance profile and phage typing of the Salmonella Enteritidis isolates. Brazilian Journal of Microbiology, v. 38, n. 2, p. 296–299, abr. 2007. Available from: https://doi.org/10.1590/S1517-83822007000200021. Acessed in: 02 mar. 2021.
SERENO, M. et al. Antimicrobial Susceptibility and Biofilm Production by Salmonella sp. Strains Isolated from Frozen Poultry Carcasses. Brazilian Journal of Poultry Science, v. 19, n. 1, p. 103–108, jan. 2017. Available from: https://doi.org/10.1590/1806-9061-2016-0268. Acessed in: 02 mar. 2021.
SINGH, S.; SINGH, S. K.; CHOWDHURY, I.; SINGH, R. Understanding the Mechanism of Bacterial Biofilms Resistance to Antimicrobial Agents. The Open Microbiology Journal, v. 11, n. 1, p. 53–62, mai. 2017. Available from: https://doi.org/10.2174/1874285801711010053. Acessed in: 02 mar. 2021.
SIVASANKAR, C.; JHA, N. K.; GHOSH, R.; SHETTY, P. H. Anti quorum sensing and anti virulence activity of tannic acid and it’s potential to breach resistance in Salmonella enterica Typhi / Paratyphi A clinical isolates. Microbial Pathogenesis, v. 138, p. 103813, 1 jan. 2020. Available from: https://doi.org/10.1016/j.micpath.2019.103813. Acessed in: 02 mar. 2021.
STEPANOVIĆ, S.; VUKOVIĆ, D.; DAKIĆ, I.; SAVIĆ, B.; ŠVABIĆ-VLAHOVIĆ, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. Journal of Microbiological Methods, v. 40, n. 2, p. 175–179, 1 abr. 2000. Available from: https://doi.org/10.1016/S0167-7012(00)00122-6. Acessed in: 02 mar. 2021.
STEPANOVIĆ, S.; VUKOVIĆ, D.; HOLA, V.; DI BONAVENTURA, G.; DJUKIĆ, S.; ĆIRKOVIĆ, I.; RUZICKA, F. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica, v. 115, n. 8, p. 891–899, 2007. Available from: https://pubmed.ncbi.nlm.nih.gov/17696944/. Acessed in: 02 mar. 2021.
SUN, D. D.; MA, X. X.; HU, J.; TIAN, Y.; PANG, L.; SHANG, H.; CUI, L. Z. Epidemiological and molecular characterization of community and hospital acquired Staphylococcus aureus strains prevailing in Shenyang, Northeastern China. The Brazilian Journal of Infectious Diseases, v. 17, n. 6, p. 682–690, 1 nov. 2013. Available from: https://doi.org/10.1016/j.bjid.2013.02.007. Acessed in: 02 mar. 2021.
THRELFALL, E. J. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiology Reviews, v. 26, n. 2, p. 141–148, 1 jun. 2002. Available from: https://academic.oup.com/femsre/article/26/2/141/653178. Acessed in: 02 mar. 2021.
VERSALOVIC, J.; KOEUTH, T.; LUPSKI, R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Research, v. 19, n. 24, p. 6823–6831, dez. 1991. Available from: https://doi.org/10.1093/nar/19.24.6823. Acessed in: 02 mar. 2021.
VOSS-RECH, D.; VAZ, C. S. L.; ALVES, L.; COLDEBELLA, A.; LEAO, J. A.; RODRIGUES, D. P.; BACK, A. A temporal study of Salmonella enterica serotypes from broiler farms in Brazil. Poultry Science, v. 94, n. 3, p. 433–441, 1 mar. 2015. Available from: https://doi.org/10.3382/ps/peu081. Acessed in: 02 mar. 2021.
VON RÜCKERT, D. A. S. et al. Pontos críticos de controle de Salmonella spp. no abate de frangos. Arquivo Brasileiro de Medicina Veterinária e Zootecnia, v. 61, n. Arq. Bras. Med. Vet. Zootec., vol. 61, n. 2, p. 326–330, abr. 2009. Available from: https://doi.org/10.1590/S0102-09352009000200007. Acessed in: 02 mar. 2021.
WANG, M.; GUO, Q.; XU, X.; WANG, X.; YE, X.; WU, S.; HOOPER, D. C.; WANG, M. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrobial Agents and Chemotherapy, v. 53, n. 5, p. 1892–1897, maio 2009. Available from: https://doi.org/10.1128/AAC.01400-08. Acessed in: 02 mar. 2021.
YIN, X.; M’IKANATHA, N. M.; NYIRABAHIZI, E.; MCDERMOTT, P. F.; TATE, H. Antimicrobial resistance in non-Typhoidal Salmonella from retail poultry meat by antibiotic usage-related production claims – United States, 2008–2017. International Journal of Food Microbiology, v. 342, p. 109044, 16 mar. 2021. Available from: https://doi.org/10.1016/j.ijfoodmicro.2021.109044. Acessed in: 02 mar. 2021.
ZIECH, R. E. Caracterização de Salmonella sp. isolada de indústrias de aves baseada na formação de biofilmes, tolerância a sanitizantes e resistência a antimicrobianos. Dissertação (Mestrado em Ciência Animal) – Programa de Pós-Graduação em Ciência Animal, Universidade Federal do Paraná. Palotina, 2015.
[1] Doctoral student of the Graduate Program in Animal Bioscience. ORCID: 0000-0003-0698-6125. CURRÍCULO LATTES: http://lattes.cnpq.br/5170743479462841.
[2] Doctoral student of the Graduate Program in Animal Bioscience. ORCID: 0000-0001-9410-7631. CURRÍCULO LATTES: http://lattes.cnpq.br/4532231119888940.
[3] PhD in Genetics. ORCID: 0000-0003-0698-6125. CURRÍCULO LATTES: http://lattes.cnpq.br/4891800920829895.
[4] Graduate in Veterinary Medicine. ORCID: 0000-0001-9273-5204. CURRÍCULO LATTES: http://lattes.cnpq.br/8329028352662293.
[5] M.Sc. in Animal Bioscience. ORCID: 0000-0002-4913-8313. CURRÍCULO LATTES: http://lattes.cnpq.br/4967459162060058.
[6] Master in Applied Cellular and Molecular Biology. ORCID: 0000-0001-7764-0573. CURRÍCULO LATTES: http://lattes.cnpq.br/6281410502740086.
[7] Bachelor of Biological Sciences. ORCID: 0000-0001-7928-7435.
CURRÍCULO LATTES: http://lattes.cnpq.br/9549169192404311.
[8] PhD in Biochemistry and Physiology, Master in Physiology, Biologist. ORCID: 0000-0003-1493-7964. CURRÍCULO LATTES: http://lattes.cnpq.br/9044747136928972.
[9] Doctor. ORCID: 0000-0002-1993-0350.
[10] Advisor. Doctor of the Graduate Program in Animal Bioscience. ORCID: 0000-0002-1289-2902. CURRÍCULO LATTES: http://lattes.cnpq.br/5998863169551704.
Sent: March 27, 2023.
Approved: April 08, 2023.