ORIGINAL ARTICLE
MIGUES, Vitor Hugo [1], AFFONSO, Paulo Roberto Antunes de Mello [2], WALDSCHMIDT, Ana Maria [3], FRANCISCO, Ana Karina [4], YAMAKI, Regina Terumi [5]
MIGUES, Vitor Hugo. Et al. An efficient method to recycle and recover silver nitrate for genetic analyses. Literature review. Revista Científica Multidisciplinar Núcleo do Conhecimento. Year 03, Issue 09, Volume 10, pp. 155-167 September 2018. ISSN: 2448-0959
ABSTRACT
Silver nitrate (AgNO3) is a routine reagent in Laboratories of Molecular Genetics and Cytogenetics, thereby resulting in large amounts of residues. The goal of this work was to establish the reaction kinetics and to determine the purity level of silver nitrate residues based on reduction–oxidation method to be reused in genetic analyses. We obtained 0.1687 g and 0.1585 g of AgNO3 after four hours of reaction with copper and iron, respectively. The purity of the recovered silver nitrate was high, reaching 86.25% (iron) and 95.98% (copper), being perfectly applicable to analyses of DNA fragments and location of nucleolar organizer regions (NORs) on chromosomes. These results show that the reduction–oxidation strategy is a viable, cost-effective, and fast method to recycle silver nitrate residues in Laboratories of Genetics.
Keywords: Genetics, NORs, Redox, Silver nitrate, Sustainable development.
INTRODUCTION
Over the last years, the disposal of residues into natural environments has been a worldwide concern for chemical industries, and both private and public research institutions1. Whenever possible, efforts should be directed to recover these residues using different methods in order to allow their reutilization3-8. This initiative is in agreement with the sustainable development by minimizing both environmental damages and risks to public health.
Since educational facilities, such as universities, play a crucial role in assuring the formation of citizens, their course of action should bring awareness to issues related to the production and disposal of residues. Nonetheless, many laboratorial activities related to experimental classes or research projects produce large amounts of harmful residues to both human health and biodiversity1.
In the case of Genetics laboratories, a solution of silver nitrate (usually at 0.2%) is commonly used to reveal DNA fragments in polyacrylamide gel after electrophoresis. Afterwards, the gel is dried and documented for analyses of genetic polymorphism while the silver nitrate solution is discarded. Furthermore, a concentrated silver nitrate solution (50%) is also employed to detect active ribosomal cistrons or nucleolar organizer regions (NORs) on chromosome spreads, followed by washing the solution in tap water. This Ag-NOR procedure is largely used in cytogenetic analyses of animals and plants to discriminate species and/or populations.²
On the other hand, high levels of silver in water, particularly ionic forms, are quite harmful to aquatic organisms. For instance, the silver ions precipitate when in contact with the mucus of gills leading fishes to asphyxia. Even though most of silver found in rivers and lakes derive from natural runoffs, high concentration of this metal are usually related to human activities. Indeed, the improper disposal of residues by laboratory activities in universities combined with the lack of efficient monitoring (at least, in low-middle-incoming countries) has been responsible for environmental degradation, besides increasing the waste of materials, what represents an ethical paradox for educational institutions.4-5
Therefore, the goal of this study was to recover silver nitrate residues from the Laboratory of Molecular Genetics and Cytogenetics from Universidade Estadual do Sudoeste da Bahia, in Jequié, BA, northeastern Brazil, based on redox reaction with copper and iron. Moreover, the recycling of silver nitrate was verified by using the recovered silver nitrate solution to stain polyacrylamide gels and locate NORs on metaphase spreads.
MATERIAL AND METHODS
The silver nitrate solution is used on a daily basis to stain DNA fragments in polyacrylamide gels and to reveals NORs on chromosomes, thus resulting in large amounts of residues after each laboratorial procedure. When untreated, or improperly discarded, the silver nitrate will reach natural environmental, being potentially harmful. Therefore, the silver residues used in the Laboratory of Molecular Genetics and Cytogenetics at UESB were recovered using redox method, quantified and applied in genetic analyses.
The redox method used in the present work was performed by immersing copper wires or iron nails into the silver nitrate solution. Once the surface contact area influences the reaction kinetics, the time of reaction is an extremely important parameter to be considered. Therefore, five reaction systems were tested, by exposing the silver nitrate solution to copper or iron for 1, 2, 3, 4, and 5 hours. After each interval, the silver on copper wire and iron nails was abundantly washed in deionized water, filtrated and dried for 30 minutes at 120ºC. Afterwards, the amount of recovered silver in solid state was weighed in analytical scale.
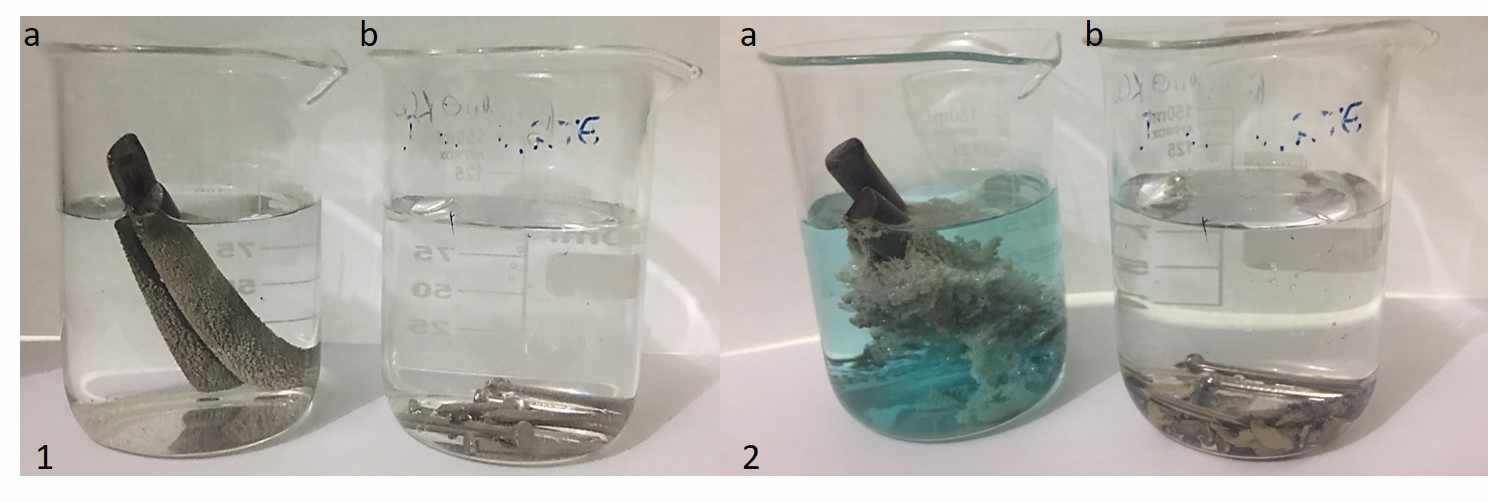
At a microscopic level, the Ag+ ions establish a direct contact with the copper and/or iron surface, where the electron transference takes place9. Two electrons from a Cu or Fe atom are transferred to two Ag+ atoms. The Cu2+ and Fe2+ ions then enter the solution and silver atoms precipitate onto the copper surface (Figure 1). This product-favored reaction goes on until one of both reagents have been entirely consumed10.
Figure 1: Redox system to recover Ag+ in silver nitrate solution (100.00 mL) using copper (a) and iron (b) at distinct periods: time zero (1) and 4 h later (2).
During the interaction between Ag+ ions and copper, Cu(0) is oxidized to Cu(II) by the transference of electrons from the copper to silver, which is reduced from Ag(I) to Ag(0), as follows:
2Ag+(aq) + 2e– 2Ag(s) E° +0,799 V
Cu2+(aq) + 2e– Cu(s) E° + 0,337 V
2Ag+(aq) + Cu(s) 2Ag(s) + Cu2+(aq) Et° +0,462 V
Likewise, when Ag+ ions interacts with iron, the Fe(0) oxidizes to Fe(II) while Ag(I) reduces to Ag(0) because electrons from iron atoms are transferred to silver:
2Ag+(aq) + 2e– 2Ag(s) E° +0,799 V
Fe(s) Fe2+(aq) + 2e– E° + 0,44 V
2Ag+(aq) + Fe(s) 2Ag(s) + Fe2+(aq) Et° +0,359 V
The spontaneity of redox reactions are predicted by the thermodynamics and entropy forces, usually expressed by the Gibbs free energy potential (ΔG°) generated by an electrochemical cell. In the case of Ag/Fe and Ag/Cu reactions, the ΔG° values equal -69.27 kJ mol–1 and -89.15 kJ mol–1, respectively. These negative values indicate a strong tendency of electron transference from the metallic Fe and Cu to Ag+ ions. It should be pointed out that this tendency in redox reaction depends only on the nature, state, and concentration of reagents and products.10
3.1 RESULTS AND DISCUSSION
REACTION KINETICS OF TEST SYSTEMS
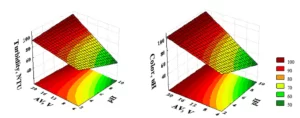
The reaction kinetics reflects the processing rate of a reaction with regards to their products or reagent depletion. In the present study, the reaction kinetics is measured by the amount of solid silver obtained in each redox system (Cu or Fe) at distinct periods (Figure 2). Even though the analysis after each test system showed the silver was the mostly recovered substance, traces of iron and copper were also detected, once these metals were lixiviated during washes.
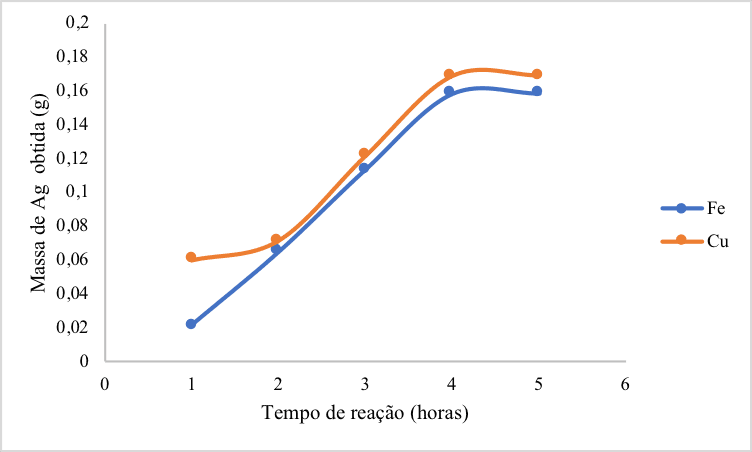
Figure 2: Graph showing the reaction time (in hours) in relation to the recovered silver mass (g) after contact with copper and iron.
Based on 300.00 mL of AgNO3 residues, the best results were observed after 4 hours of reaction, since shorter periods resulted in low rates of recovered material while longer periods presented constant recovery rates. In fact, the reaction time was relatively long because of the large area of selected electrodes. A slight difference in the silver mass was observed after 4 hours depending on the tested metal, being equal to 0.1687 g using copper and 0.1585 g using iron. Thus, the copper reaction was more efficient in recovering larger amounts of silver than iron (Figure 2), confirming the high redox Cu/Ag potential, as previously indicated by ΔG° values.
The yield of silver recovery could not be precisely estimated based on initial concentrations because the silver nitrate residues were previously used to stain polyacrylamide gels and chromosomes. Therefore, we carried out a control reaction based on the recovery of 2.0002 g of pure silver nitrate (VETEC PA). As a result, we verified that the tested method was successful, being able to recover 94% (1.8819 g) of the initial silver mass.
3.2 RECOVERY AND PURITY OF SILVER NITRATE
In order to test the reutilization of recovered silver in genetic analysis, we added stoichiometrically calculated amounts of nitric acid (HNO3 65% v/v VETEC) to the obtained silver mass in each tested system (0.5226 g and 0.5782 g of Ag using iron and copper, respectively). Thus, a solution of silver nitrate was produced, as follows:
Ag(s) + 2HNO3(aq) NO2(g) + H2O(l) + AgNO3(aq)
The silver nitrate solution was then heated to evaporate the excess of nitric acid and the water, followed by immersion in iced bath to crystallization. Right after, the crystals were filtrated and dried in heated chamber.
The purity of the recovered silver nitrate was determined according to Mohr’s method.11-13 This argentimetric method is applicable to determine chloride (Cl–). Such procedure is based on the precipitation titration of chloride ions in the presence of silver nitrate. For that, a solution of sodium chloride is titrated with AgNO3 and K2CrO4 (indicator). The end point is determined by the precipitation of red particles as result of the reaction between Ag and the indicator, as shown below:
Ag+ + Cl– AgCl(s)
2Ag+ + CrO42- Ag2CrO4(s)
A silver nitrate solution can be used as a secondary pattern to the determination of chloride, in the presence of sodium chloride. Here, a silver nitrate mass of 0.2800 g was used for the standardization to a final volume of 250.00 mL of NaCl. Then, aliquots of 25.00 mL of NaCl solution (in five replicates) were compared to a commercial sample of silver nitrate (VETEC) with 99% of purity. This approach allowed estimating the titration equivalent volumes of the recovered silver nitrate (table 1).
Table 1: Volume of NaCl (0.09541 mol.L-1) used in the volumetric titration.
| Solution | Volume NaCl (mL) |
| AgNO3 (VETEC) | 5.02 ± 0.03 |
| AgNO3 (recovered with iron) | 5.23 ± 0.03 |
| AgNO3 (recovered with copper) | 5.82 ± 0.03 |
The mean volume of silver nitrate solution used in the control analysis was 5.02 mL, while the silver nitrate volume after recovery using copper and iron were 5.23 mL and 5.82 mL, respectively. Thus, the purity of recovered AgNO3 varied from 86.25% (iron system) and 95.98% (copper system), showing a better performance of the latter.
Even though the reduction of Ag ions in the presence of Cu might produce harmful residues to the environment, they can be easily recovered in the form of copper sulphate33 in a simple reaction between Cu ions, sodium hydroxide and diluted sulfuric acid.
3.3 APPLICABILITY OF RECOVERED SILVER NITRATE
3.3.1 STAINING OF POLYACRYLAMIDE GELS
Polyacrylamide gels are widely used to separate small DNA fragments (<1,000 base pairs) after electrophoresis, being more sensitive than agarose gels to genetic analysis such as genotyping based on the differential size of repetitive sequences.14 Nonetheless, the visualization of DNA fragment in polyacrylamide gels depends on the staining using silver nitrate solution. In the present study, polyacrylamide gels containing DNA fragments of microsatellite loci (short in tandem repeats) were stained as determined by Bassem et al.15 using pure silver nitrate solution (control) and the recovered silver nitrate (copper and iron-based systems), as shown in Figure 3.
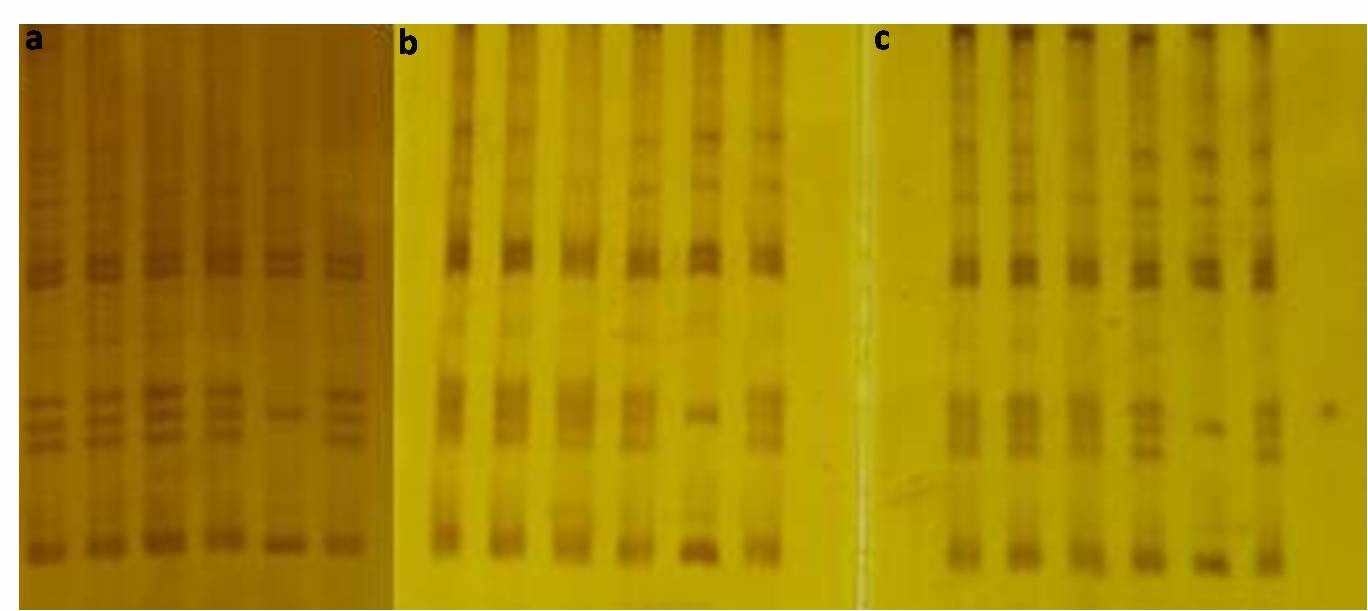
Figure 3: Polyacrylamide gel stained with pure (a) and recovered silver nitrate in iron (b) and copper (c) systems, revealing the profile of DNA bands after amplification of microsatellite loci from a bee species of the genus Melipona.
In spite of the slight differences in the staining intensity in each gel, the DNA fragments were invariably detectable using the three distinct silver nitrate solutions. Most likely, these differences are related to the purity of recovered silver in each test (99.99% for pure AgNO3, 95.98% in copper-based system, and 86.25% in the iron-based system). These results show that the recovered silver nitrate can be used routinely in molecular genetics laboratories, decreasing the productions of contaminant residues.
3.3.2 MAPPING OF AG-NORS ON CHROMOSOMES
The nucleolar organizer regions (NORs) comprise the genes encoding ribosomal RNA (rRNA) of the 45S subunit. Active NORs show high affinities with silver ions by the presence of non-histones proteins associated to rRNA.15 Therefore, the silver nitrate staining as developed by Howell and Black16 is a fast and simple method to reveal active NORs on chromosomes, being named as Ag-NOR staining. We used the same methodology to reveal NORs by staining the metaphase spreads obtained according to Foresti et al.17 from a tropical fish species (Metynnis maculatus) with pure (control) and recovered silver nitrate solutions at 50% (Figure 4).
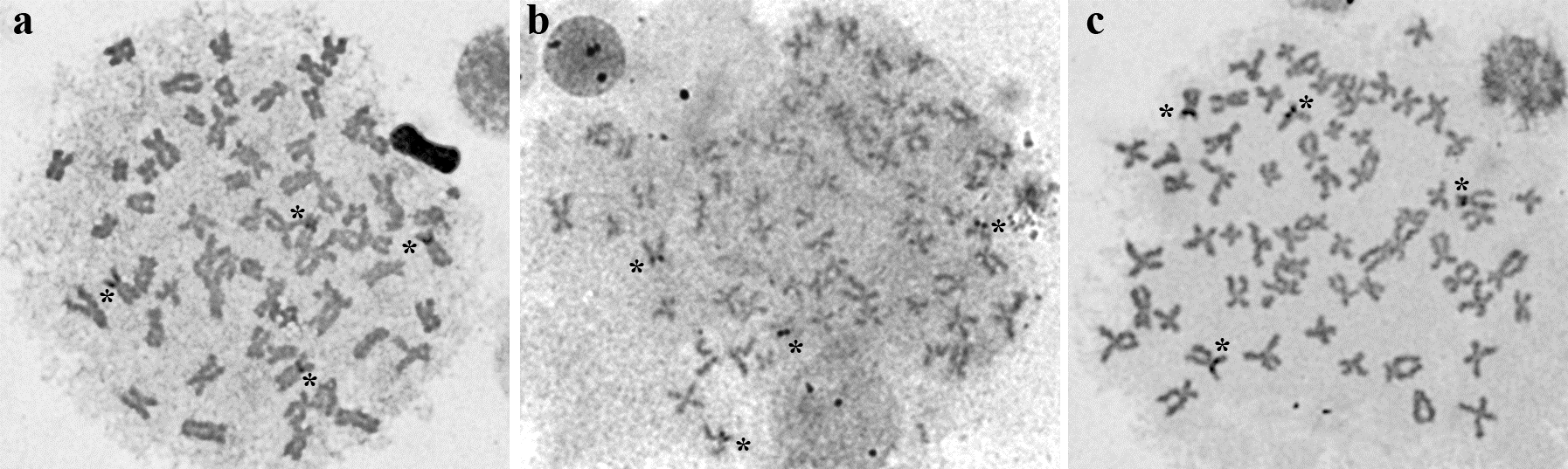
Figure 4: Metaphases of M. maculatus after silver nitrate staining using pure silver (a) and recovered silver nitrate based on iron (b) and copper (c) reactions. The stars indicate the active NORs on four chromosomes.
Again, both pure and recovered silver nitrate solutions revealed the same pattern of Ag-NOR staining on chromosomes. Nonetheless, the quality of metaphases stained with the silver nitrate after recovery of Ag ions using copper was higher than that observed using iron (Figure 4), probably because of the higher purity of the former, as pointed out in polyacrylamide gels.
4. CONCLUSION
The present data reveal that a simple procedure to recover silver nitrate can be successfully used to routine analysis of cytogenetics and molecular biology, particularly based on redox reaction with copper. This procedure represents a significant improvement to reduce the amount of residues in laboratories of genetics. Furthermore, we encourage other scientists and technicians to test the recovered silver nitrate in other laboratorial experiments to evaluate the extension of its applicability.
Considering the increased levels of pollution worldwide, efficient measurements to reduce the environmental contamination should be promoted38, as established by the Kyoto’s protocol18. In the case of University laboratories, the incorporation of the reutilization of residues plays an additional educational role by bringing knowledge and awareness to the students so they can spread these practices in their professional careers19. Besides being ecologically friendly, the development of recycling procedures as herein presented also reduces the costs in research projects and experimental classes and optimizes the financial resources.
REFERENCES
1. Amaral, S. T.; Machado, P. F. L.; Peralba, M. C. R.; Camara, M. R.; Santos, T.; Berleze, A. L.; Falcão, H. L.; Martinelli, M.; Gonçalves, R. S.; Oliveira, E. R.; Brasil, J. L.; Araújo. M. A.; Borges, A. C. A. Relato de uma experiência: recuperação e cadastramento de resíduos dos laboratórios de graduação do instituto de química da universidade federal do rio grande do sul. Química Nova 2001, 24, 419. [CrossRef]
2. Ferreira, M. E.; Grattapaglia, D. Introdução ao uso de marcadores moleculares em análise genética. 2 ed. Brasília: Embrapa-Cenargen, 1998. 220 p
3. Ishibasshi, Y.; Arizone, K.; Trials for Recyclings of the Waste Containing Heavy Metals. Journal of Health Science 1999, 45, 87. [CrossReef]
4. Kothari, A. C. Recovery of ammonia from effluents. Chemical engineering world 1999, 34,101. [CrossReef]
5. Mohanty, S. P.; Senapati, M. R. Effect of group velocity dispersion on a WDM optical ring network. Lasers and Electro-Optics Society Annual Meeting, 1998, 2, 75. [CrossReef]
6. Caabeiro, J. C. Utilization of waste water dairy sludges, Alimentaria 1999, 302, 73. [CrossReef]
7. Mishra, S. K. Resource recovery in waste treatment increasingly used. Mining engineering 1999, 51, 29. [CrossReef]
8. Zhang, Y.; Muzzy, J. D. Recycling carpet waste by injection and compression molding. Polymer-Plastics Technology and Engineering 1999, 38, 485. [CrossReef]
9. Russell, J. B. Quimica Geral – Vol. 2 Editora: Pearson Brasil. 1994.
10. Kotz, J. C.; Treichel, P. M.; Weaver, G. C. Química Geral e Reações Químicas. Vol. 2. Cengage Learning. 2009.
11. Vogel, A. I. Análise Química Quantitativa. 6.ed. Rio de Janeiro: LTC-Livros Técnicos e Científicos, 2002. 462 p.
12. Skoog, A. D.; West, D. M.; Holler, F. J.; Fundamentals of Analytical Chemistry, 7a ed., Saunders College Publishing: Orlando, 1997.
13. Baccan, N.; Andrade, J. C.; Godinho, O. E. S.; Barone, J. S. Química Analítica Quantitativa Elementar. 3.ed. São Paulo: Edgard Blucher, 2000.
14. Bassam, B. J.; Caetano-Anollés, G.; Gresshoff, P. M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Analytical Biochemistry 1991, 196, 80. [PubMed]
15. Faria, B. A.; Oliveira, S. M.; Santos, A. P. Enciclopédia Biosfera, Centro Científico Conhecer. 2010.
16. Howell, W. M.; Black, D. A. 1980. Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia 1980. 36, 1014. [Pubmed]
17. Foresti, F.; Oliveira, C.; Almeida-Toledo, L. F. A method for chromosome preparations from large specimens of fishes using in vitro short treatment with colchicine. Experientia 1993, 49, 810. [CrossReef]
18. “Protocolo de Kioto” 2002. [Link]
19. Cunha, C. J. O programa de gerenciamento dos resíduos laboratoriais do depto de química da UFPR. Química Nova 2001, 24, 424. [CrossReef]
[1] He holds a Bachelor’s degree in Chemistry from the State University of Southwest of Bahia (2012) and a Master’s Degree in Analytical Chemistry (2014) from the State University of Southwest of Bahia. She is currently a doctoral student in Organic Chemistry at the Federal University of Bahia. He has experience in the area of determination of metallic ions in the most diverse matrices by UV-Vis, ICP OES, FAAS and ICP-MS.
[2] He holds a degree in Veterinary Medicine from the Fluminense Federal University (1997), a Masters in Genetics and Evolution from the Federal University of São Carlos (2000) and a PhD in Genetics and Evolution from the Federal University of São Carlos (2004). He is a full professor at the State University of Southwest of Bahia (Department of Biological Sciences) and permanent advisor of the Postgraduate Program in Genetics, Biodiversity and Conservation of UESB in Jequié-BA, acting as coordinator from 2014 to 2016 and vice-coordinator since July 2018. He also works in the Postgraduate Program in Biodiversity and Evolution (formally Animal Diversity) of the Federal University of Bahia – UFBA.
[3] Federal University of Uberlândia (1988), Masters in Genetics and Improvement by the Federal University of Viçosa (1995) and PhD in Genetics and Improvement by the Federal University of Viçosa (1999). Currently holds a teaching position in the Plena category at the State University of Southwest of Bahia (UESB). She is coordinator of the molecular biology laboratory at UESB, Jequié campus. She is the coordinator of the Postgraduate Program in Genetics Biodiversity and Conservation (2018-2020).
[4] He holds a degree in Biological Sciences from the Federal University of São Carlos (2000), a Masters in Genetics and Evolution (2003) and a PhD in Sciences, with an emphasis on Genetics and Evolution (2009) by the same University. She is currently working as an Adjunct Professor of the Biological Sciences Department of the State University of Southwest Bahia (UESB). Has experience in the field of Animal Genetics, with emphasis in Molecula Biology.
[5] He has a degree in Pharmacy from the Federal University of Ouro Preto (1988), a Masters in Chemistry from the Federal University of Minas Gerais (1992) and a PhD in Chemistry from the Federal University of Minas Gerais (1999). He is currently an associate at the State University of Southwest of Bahia. Has experience in the area of Chemical Equilibrium of Bioinorganic compounds, currently works with the study of Chemical Equilibrium of azocorantes compounds by spectrophotometry and potenciometry and application of the azocorantes compounds in systems of pre-concentration of metals.
Received: December 2017
Approved: September 2018